M-tortuosity (2D/3D)
- scalable topological descriptor; informations of different dimensions
random points sampling, handling disconnections and applicable on complex multi-scale microstructures, especially when entries and exits are delicate to impose
- the M-tortuosity is translation, rotation and homothety invariant. Moreover, it is stable for periodic microstructures.
- Optional step: skeletonization
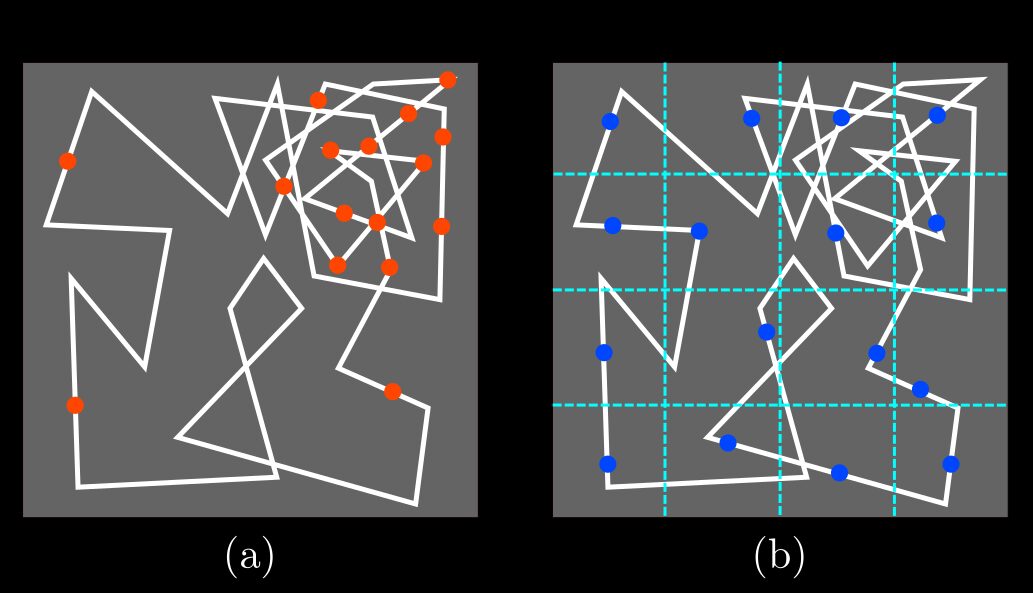
- Sampling: definition of the set of N random points; 1D-sampling method (a), stratified-sampling method
(b).
Fig.(a-b)
displays the difference between the two sampling methods.
- Distance transforms: At this step, the map of mean relative tortuosities and the maps associated to each points, if 'Save Data' checked, are computed.
- M-coefficients:
set of mean morphological tortuosities, each one connected to a
specific location, i.e. a random point.
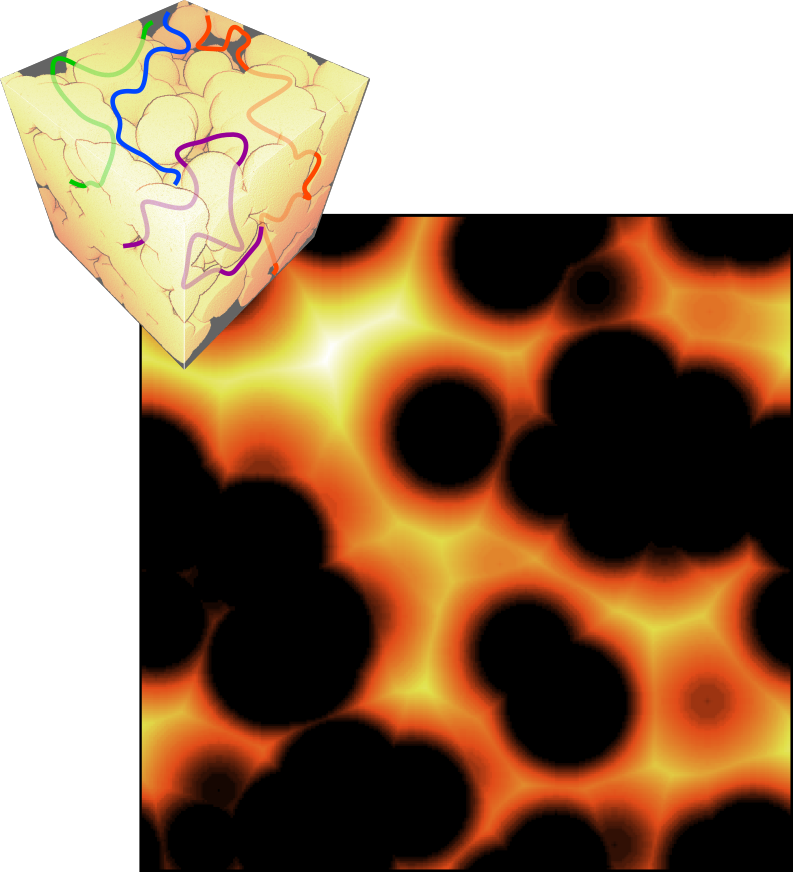
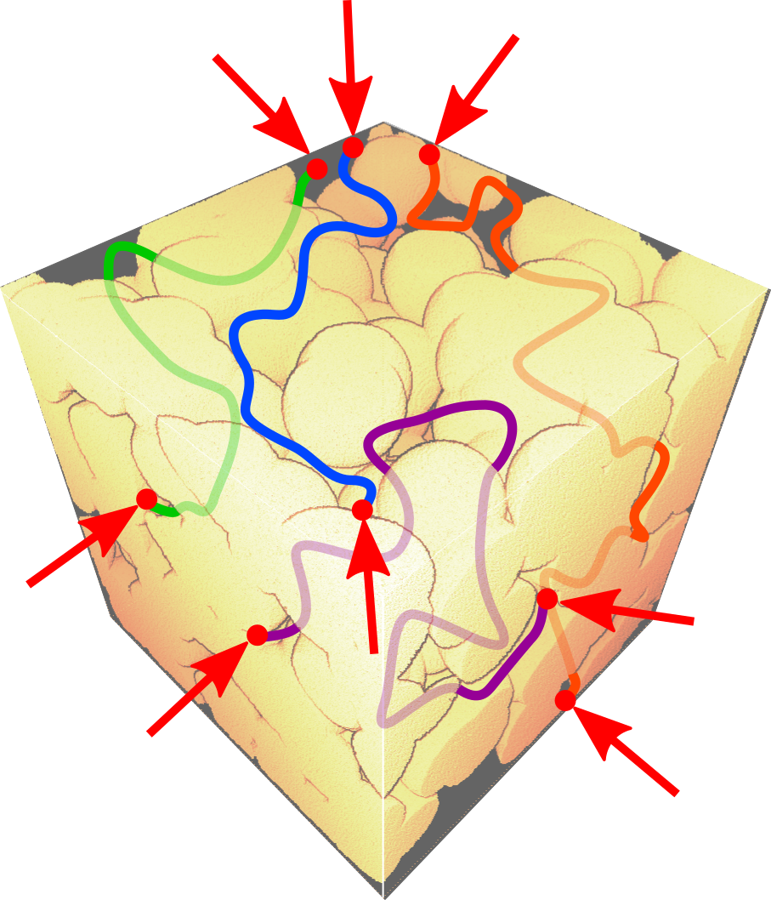
A M-coefficient can be seen as either a viewpoint of the microstructure from this location, or the mean accessibility of this point for any random point. - M-scalar:
The M-tortuosity value, named the M-scalar, is an assessment of the morphological tortuosity. It aggregates information of each M-coefficient into a scalar value.
The M-scalar is a global viewpoint of the microstructure, or the mean accessibility of any random point for any other random point.
- binary microstructure
Outputs:
- M-tortuosity value
(*.txt file)
- set of M-coefficients
(*.txt file)
- map of the mean relative tortuosities (*.fda file)
- optional: "Euclidean distance/geodesic distance/ morphological tortuosity" for each points pair (*.txt file)
- optional: maps associated to each starting point (*.fda file)
Parameters:
- sampling choice (1D-sampling or stratified-sampling)
- number of random points N
- boolean 'Save Dist/Tor' (optional output): save information for each points pair; distances and tortuosity
(*.txt file)
- boolean 'Save Data' (optional output): maps associated to each starting point (*.fda file) . Additional parameters (if checked):
- 'Maps': morphological tortuosities map of each point
- 'Paths': geodesic paths associated to each point
- '...': selection of the save directory